Miller RS, MacLean AR, Gunson RN, Carman WF. Occurrence of haemagglutinin mutation D222G in pandemic influenza A(H1N1) infected patients in the West of Scotland, United Kingdom, 2009-10. Euro Surveill. 2010;15(16):pii=19546
Since the original report [1] several countries have detected this mutation [2]. This data has been summarised in a recent World Health Organization (WHO) review, which reported that the overall prevalence of D222G was <1.8% in contrast to a rate of 7.1% in fatal cases [2]. The WHO paper also reports on the occurrence of other mutations at this amino acid, D222E and D222N, although their significance is unclear. A group in Hong Kong have also analysed this amino acid in severe and non-severe cases of pandemic influenza A(H1N1) [3]. In this study nine (4.1%) of 219 severe or fatal cases of pandemic influenza A(H1N1) had the D222G mutation, in contrast to 0 of 239 non-severe cases.
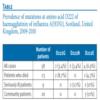
We sequenced the HA1 subunit of the HA gene from a number of West of Scotland cases, both community cases and severely ill. Furthermore we subdivided the severely ill into those who had died and those who recovered after hospitalisation. We found an increased incidence of D222G in those patients who died (2/23 - 8.7%) compared to both community and hospitalised patients (0/35 - 0%). We also detected an increased incidence (2/32 - 6.2% cf 0/26 - 0%) of D222N (aspartic acid to asparigine) in severely ill patients and those who had died. The significance of this mutation is unclear. There was a low level of D222E (aspartic acid to glutamic acid) present in both severely ill and community cases with no significant difference between the two. The results are summarised in the Table.
Table. Prevalence of mutations at amino acid D222 of haemagglutinin of A(H1N1), Scotland, United Kingdom, 2009-2010
Interestingly, in one of the patients who died and had the D222G mutation, the original sequence had a mixed base in the D222 codon giving D222D/G. On resequencing two more samples from this patient, we obtained a pure D222G on one occasion and a pure wildtype D222 on the other, showing that this patient had a mixed population of virus. This confirms the finding in Kilander’s paper [1] of the co-existence of mutant and wildtype virus.
References
- Kilander A, Rykkvin R, Dudman S, Hungnes O. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009-2010. Euro Surveill. 2010;15(9) pii=19498. Available from:http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19498
- World Health Organization. Preliminary review of D222G amino acid substitution in the haemagglutinin of pandemic influenza A(H1N1) 2009 viruses. Wkly Epidemiol Rec. 2010;85(4):21-2
- Mak GC, Au KW, Tai LS, Chuang KC, Cheng KC, Shiu TC, Lim W. Association of D222G substitution in haemagglutinin of 2009 pandemic influenza A (H1N1) with severe disease. Euro Surveill. 2010;15(14): pii=19534. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19534
See Also:
Latest articles in those days:
- Risk of infection of dairy cattle in the EU with highly pathogenic avian influenza virus affecting dairy cows in the United States of America (H5N1, Eurasian lineage goose/Guangdong clade 2.3.4.4b. ge 12 hours ago
- Avian influenza overview September - November 2025 13 hours ago
- [preprint]Airway organoids reveal patterns of Influenza A tropism and adaptation in wildlife species 13 hours ago
- Cats are more susceptible to the prevalent H3 subtype influenza viruses than dogs 15 hours ago
- Overview of high pathogenicity avian influenza H5N1 clade 2.3.4.4b in wildlife from Central and South America, October 2022-September 2025 15 hours ago
[Go Top] [Close Window]